Ginkgo.Your Innovation Partner.

This summer, Ginkgo welcomed six interns to the Digital Technology Team. These six Software Engineering and Product Management interns came to us with diverse backgrounds from universities all across the US. Each intern completed a project that leverages cutting-edge technologies such as React, GraphQL, Django, Airflow, Docker, and AWS to accelerate and scale Ginkgo’s mission to make biology easier to engineer.
Due to the pandemic we were not able to welcome them to our ocean-side office in person. However, our interns were able to integrate into our scrum teams and get to know their mentors and teammates through remote conferencing and collaboration technologies such as Zoom and Slack. They heard tech and career talks from Ginkgo and Digital Technology leaders, joined seminars given by our scientists and biological engineers, and even had virtual coffee with our CEO. With happy hours, game nights, and digital scavenger hunts, we have had a fun, productive, and fulfilling summer!
But, don’t take my word for it! Here, three of our interns will tell you about their summers in their own words.
This summer I interned on the product management team. I worked on Loom, an internal DNA design and ordering software platform. Users create a scaffold of DNA building blocks in the Loom workspace and then add designer sequences, called design units, to create full DNA constructs. Users typically hand off the design to a reviewer to check the validity of the constructs before they are ordered. Design reviews save time and money by mitigating the risk of incorrect DNA sequences. But what if the design review process could be made more efficient?
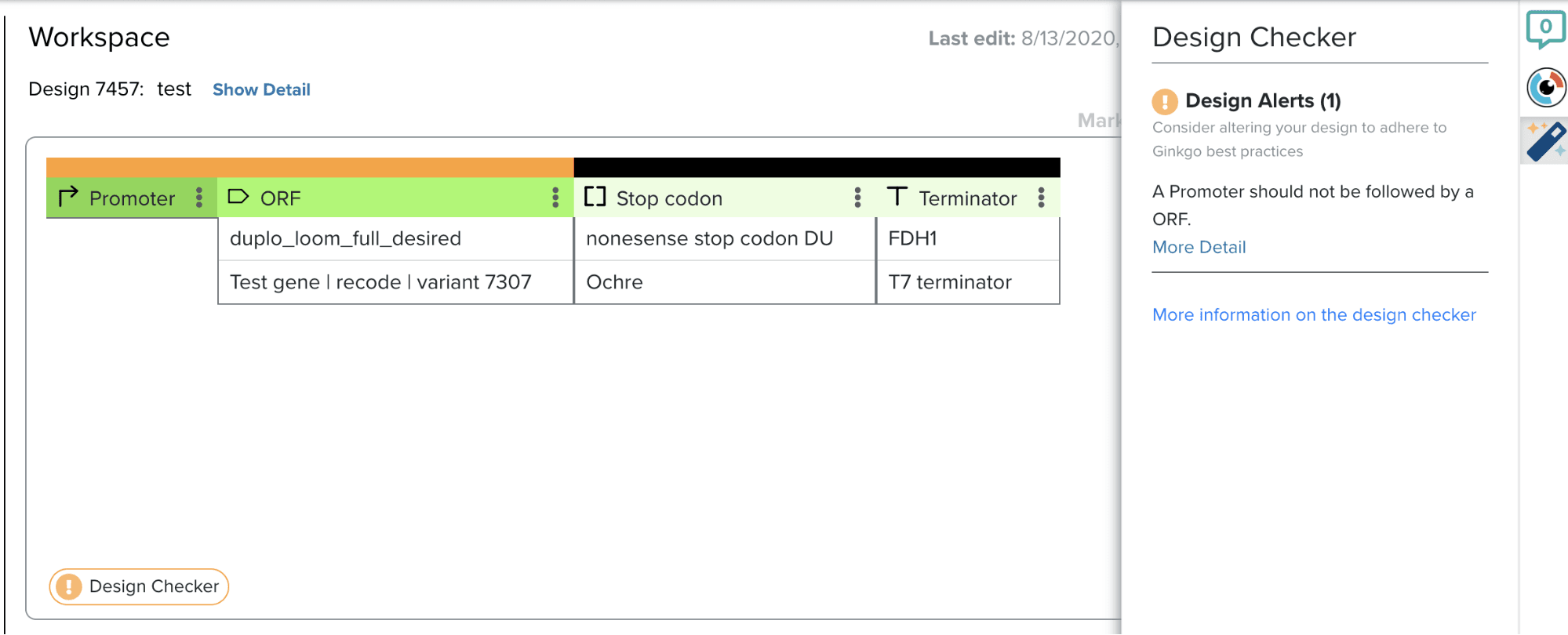
Design validations are the solution: if computational checks could be implemented at strategic points in the workflow, DNA designing could be simplified. One such feature would be an iterative design checker, providing real time feedback as the user designs in the interface. Daphne, another intern, helped build a prototype for user testing. We focused on the validation of the order of different building block types (Promoters, RBS, ORFs, etc…) and developed validators unique to prokaryotic and eukaryotic organisms. The checker provides unobtrusive alerts for errors, identifies them in the workspace via mouseover, and suggests more appropriate block types.
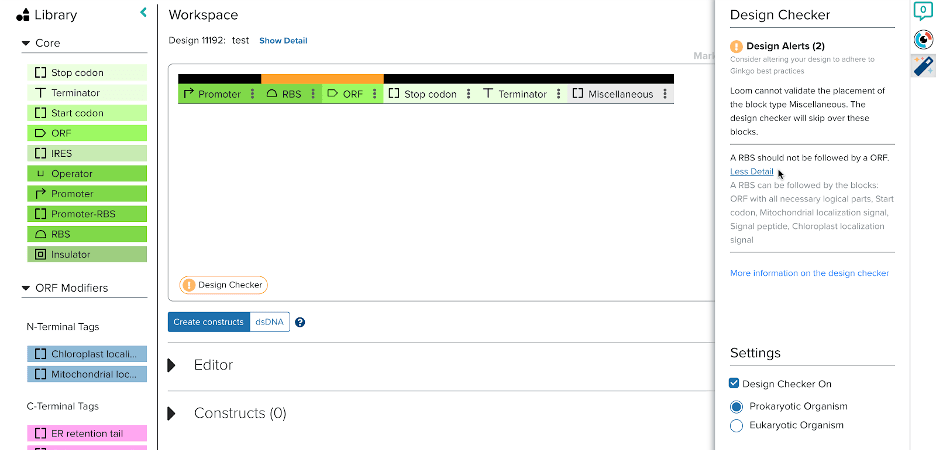
Designing DNA is not straightforward and all rules seem to have exceptions. In order to account for edge cases, the ultimate decision was left to the users’ expertise. The design checker can also be turned on and off, allowing users to check designs when they are ready. Users particularly liked that the feature did not interrupt the workflow and the ability to reference the documentation to better understand the rules. In the future, functionality for sequence level validations and for users to add their own validations could greatly improve the value of the design checker.
The ultimate vision of Loom is to democratize and scale the DNA design and ordering process. The software tool addresses this partially, but procedures and guidelines make cross-team collaboration and a reusable design unit library more attainable. One key lesson learned was that not all problems need a technical solution; alongside the software features, I compiled a centralized set of best practices on Confluence. Ideally the combination of best practices and levels of design validations will help scale Loom and increase accessibility.
One of Ginkgo’s key features is its “virtuous feedback loop” where data is generated from experiments, and then used to guide further experiments. The Software team helps facilitate that loop by enabling the flow of an immense amount of data throughout the Foundry. One way the team envisions streamlining the process is through a software platform that Ginkgo has developed called Servicely.
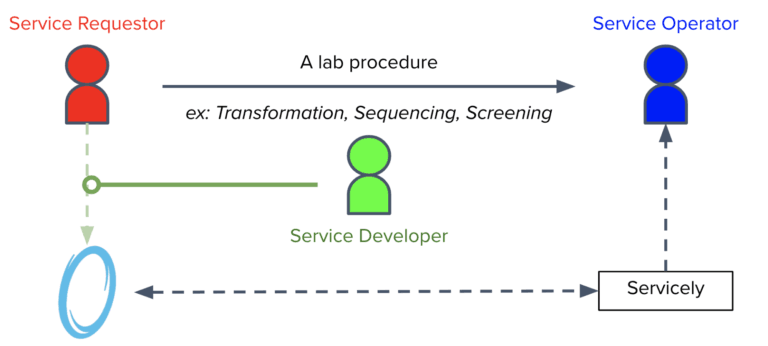
Currently, Foundry services, which represent well defined bioengineering procedures, are organized and stored in Servicely. Through this application, members of the foundry can request for lab operations to be conducted by Service Operators, which may be people or robots. The interaction between these requesters and Servicely is defined by Service Developers. While these developers have mostly been within the Software team, the JSON Schema form building tool that my internship has been focused on will help the scientists and biological engineers of the Foundry to build out the software interactions of their own services without having to code at all. In the long term, this will help Ginkgo build out and maintain more lab activities without having to expand the software team proportionately.
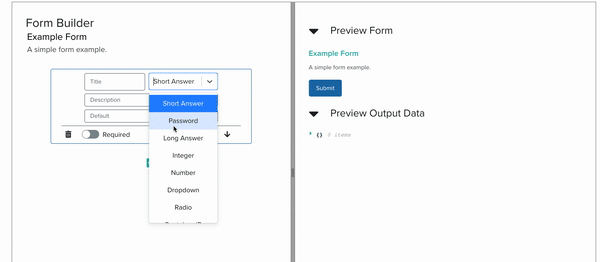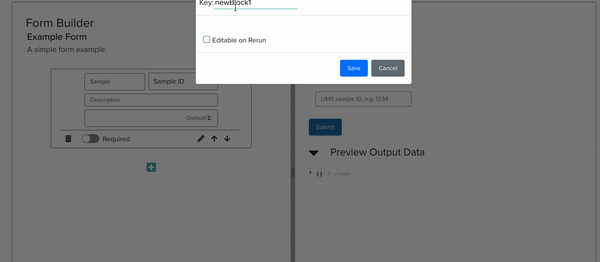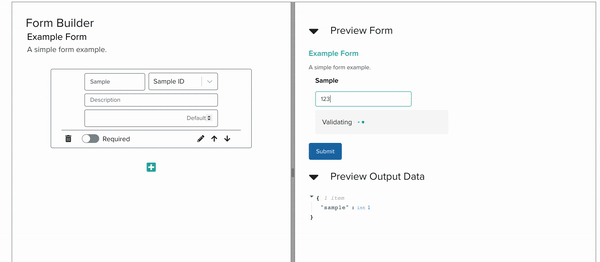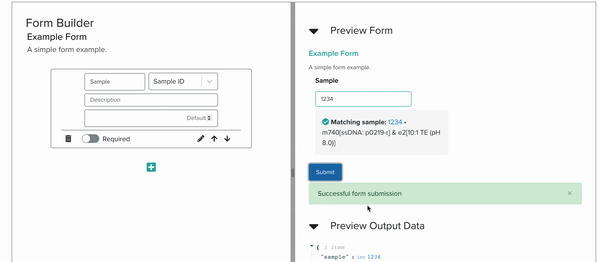
I implemented this low-code/no-code JSON Schema Form building tool by building a YAML editor, a panel with cards corresponding to form elements, and another panel which renders a preview of the form itself, all interacting with each other live. As the project progressed, we steadily approached full compatibility with JSON Schema features.
Throughout the project, I’ve learned a couple of key lessons that kept me from getting bogged down with frustrating ReactJS behavior or unpleasant UI experiences. Sticking to stateless, functional code, limiting components to ~200 lines, and keeping a separate utility file helped with unit testing and reusability, while demoing early with unfinished products helped keep the project aligned to what the users actually wanted. Finally, planning orthogonal tasks for parallel code review and development and keeping commits small were essential skills I picked up about working in a team coding environment this summer at Ginkgo!
This summer, I interned on the DNA Tools team. Our team built and maintains Loom, a software tool to enable our organism engineers to design DNA by drag-and-dropping blocks of reusable DNA parts in a web interface. While this software is great at abstracting away the complexity of synthesizing DNA and automating the ordering process, one of the opportunities to improve our tool lies within actually designing the DNA. Designing DNA takes a lot of biological knowledge. Our goal is to take the vast knowledge from our expert scientists and engineers, and build it into our software. This will enable everyone to benefit from the entirety of our in house expertise, and democratize the process of designing DNA.
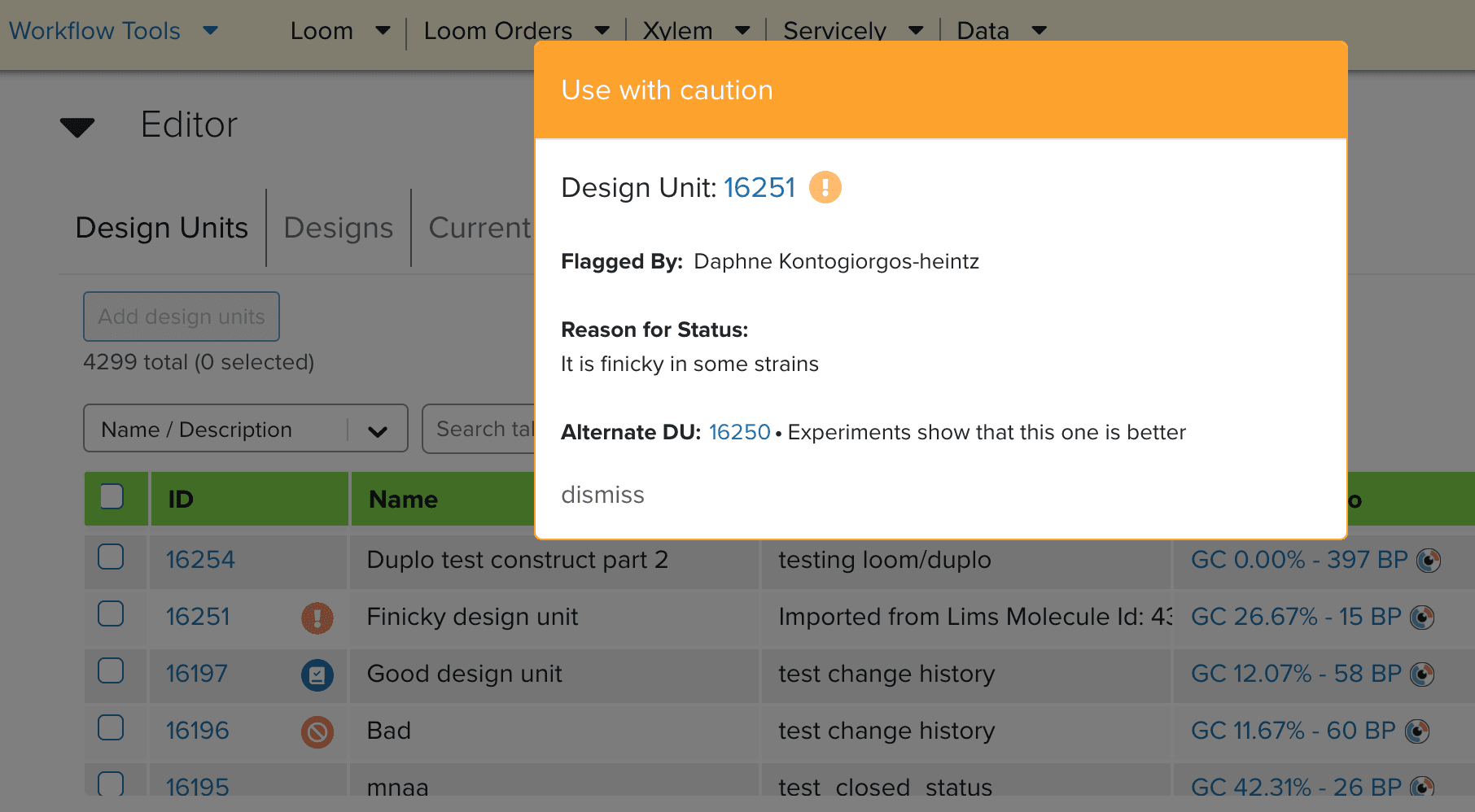
This summer, I addressed this opportunity in two ways. The first was to enable assigning “statuses” to our DNA parts. Using the entirety of our Python/Django/GraphQL/React stack, I added the new functionality for users to assign a “do not use”, “use with caution”, or “certified” status to the DNA parts. Because of the highly contextual nature of synthetic biology, these statuses, along with comments, links to alternative parts, and layers of alerts surrounding them, will help make sense of our immense library of DNA parts.
The second way I addressed this opportunity to democratize the process of designing DNA was by implementing a prototype of Rupsa’s design checker project (check out her section if you haven’t already!). One perk of creating software that is used internally is the ease of rapidly building a prototype for a new feature and getting user feedback. Building out this design checker was a lot of fun, because I got to work with one of our DNA designers to learn about what rules we should enforce, and it was fun learning more about the ways people interact with our software!
Stay tuned — in the next blog post we will be hearing about the cool things that Joseph, Addie and Vidya have been doing this summer!
(Feature photo by Ramon Kagie on Unsplash)
Posted by Rupsa Acharya, Nathan Alam, and Daphne Kontogiorgos-Heintz