Ginkgo.Your Innovation Partner.
In our last blog post, Rupsa, Nathan, and Daphne told us about the cool things that they accomplished during their summer internships with the Ginkgo Digital Tech Team. Here, we hear from software engineering interns Joseph, Addie, and Vidya!
Hello! My name is Joseph, and I interned with the Solution Engineering team this summer. I recently graduated undergrad from Caltech with a double-major in Biology and Computer Science. I am interested in the intersection of these two fields, especially in how we can apply computer science to biology in order to make sense of complex biological data. I enjoy developing software to solve challenging problems in biology, and this passion led me to Ginkgo. This is my second time working at Ginkgo, the first time was last summer when I worked on migrating Ginkgo’s next-generation sequencing (NGS) pipeline to AWS (shameless plug here and here).
This year, I was tasked with migrating pipelines running on an on-site Windows server to a more robust, scalable Linux solution. These pipelines pre-processed data generated by various laboratory instruments and uploaded them to our data store so that they could be accessed by the enduser. We had to use a Windows machine because several pre-processing software had Windows dependencies (such as .NET and Visual Studio). The current pipelines were difficult to monitor and debug, and deploys were non-standard, requiring us to remote into the host and manually copy over updates. On the surface, this seemed like a straightforward project, but it turned out to be a lot more challenging to migrate to a Linux-only solution because there were few Windows-only software with Windows DLL dependencies. For instance, the pipeline contained Windows-only software that was used to convert vendor-specific, proprietary data formats to an XML-based format called mzML. The first (and hardest) step was to containerize the conversion process into a Docker container, and this was done by emulating Windows with a software called Wine within Ubuntu.
Once this was done, it was a matter of implementation. I decided to orchestrate the pipeline on Airflow, Apache’s workflow orchestration software, because it has proven to be scalable and fault-tolerant when running Ginkgo’s NGS pipelines. The conversion itself is done in AWS Batch through an abstraction layer that we developed called Batch-as-a-Service (BaaS), which allows you to submit jobs and retrieve its results via a simple HTTP API endpoint. Here is the diagram of the new pipeline.
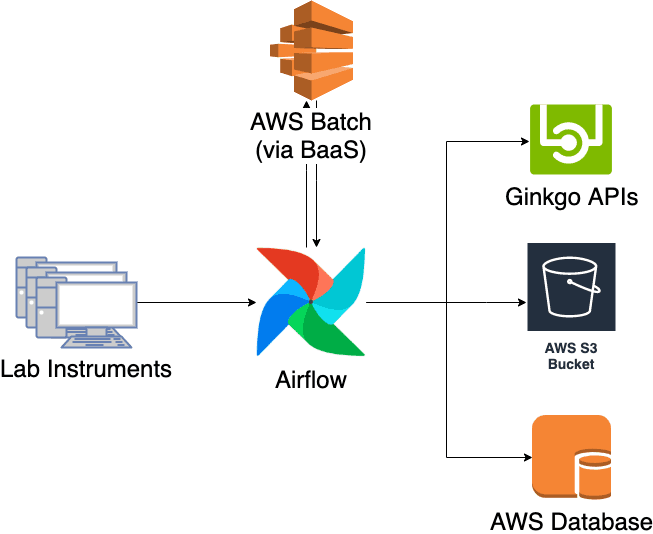
The new pipeline addresses all the challenges we were facing with the old pipeline. Testing and deployment is now tied to our standard CI/CD pipeline, allowing for a faster and more robust development cycle. Additionally, it is possible to view the pipeline status and progress in real-time through the Airflow UI, serving as a valuable resource for the software team.
I mentioned at the beginning that I worked on the Ginkgo NGS pipeline last summer. During that project, I was laser-focused on a single aspect of Ginkgo, but this year as a Solution Engineering intern I was able to experience a different, more systems-level approach to software engineering. I had a great deal of autonomy in making architectural and design decisions. Another great part (and this was also true last year) was that I got to see my project released to the rest of Ginkgo and used for live data. I think Ginkgo’s internship culture is quite unique in how much trust and responsibility Ginkgo puts on its interns, and it has personally served as great motivation and excitement, knowing that the goal of my project was to release a finished product for use by the entire company.
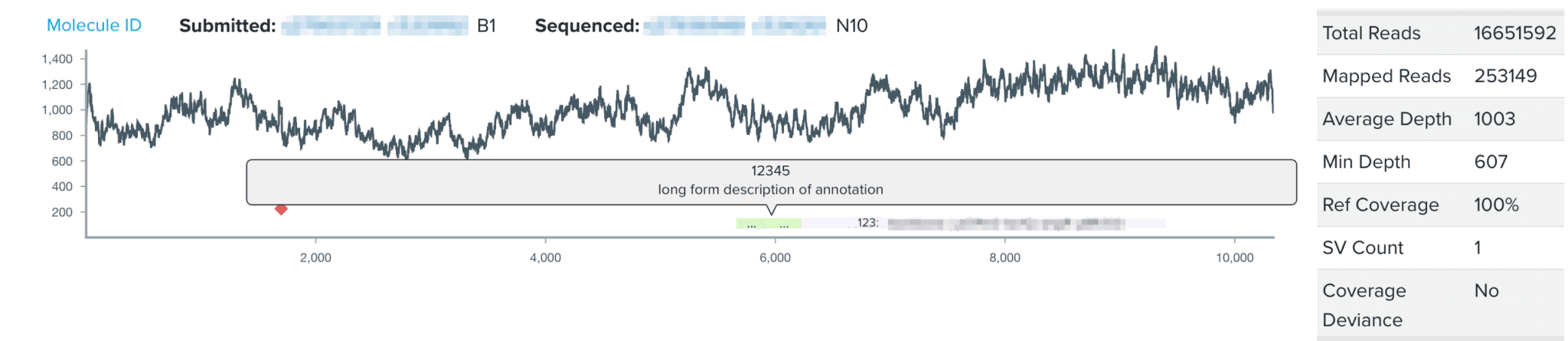
During the design phase of the design-build-test cycle, biologists add annotations to subsections of their DNA constructs, or DNA design units. Such annotations directly contribute to our goal at Ginkgo of making biology easier to engineer; by labeling subsequences of DNA with their functionality or any additional details through our DNA design tool, biologists can easily reuse DNA parts and collaborate with other biologists’ previously constructed DNA parts. However, the NGS sequence viewer, where biologists analyze and validate the sequence of their DNA construct, did not visualize the DNA design unit annotations users had created during the design stage. For my summer project, I have worked on integrating users’ design unit annotations from our DNA design tool into our sequence viewer for a more seamless user experience. By visualizing annotations with increased granularity in the sequence viewer, this project aims to increase the efficiency of biologists’ sequence validation, encourage high-quality annotations, and work to connect software tools across the design-build-test cycle of synthetic biology.
Throughout my internship, I completed an end-to-end design process, beginning by interviewing organism engineers to identify key pain points for users analyzing sequencing data with our sequence viewer. We then developed the user experience vision for the project, where I collaborated across software teams to understand technical details of the DNA design tool, consulted with UX designers and product managers to develop the project vision and to plan out project validation and execution. In the project implementation, I utilized our Python/Django backend to generate BED files for each annotation source, and developed GraphQL queries to accommodate multiple annotation types. To finally realize the vision cultivated by the team, I implemented the visualization of the DNA design unit annotations in our sequence viewer in a React framework.
In addition to the abundance of technical knowledge and growth I have developed this summer at Ginkgo, the team at Ginkgo has also taught me practical skills to become a better engineer and teammate. I have learned to question my own initial assumptions, which has led me to write more robust code with better error handling and ask better questions. I have also learned to communicate effectively with different stakeholders to achieve a unified design concept, a key step in developing software for biologists.
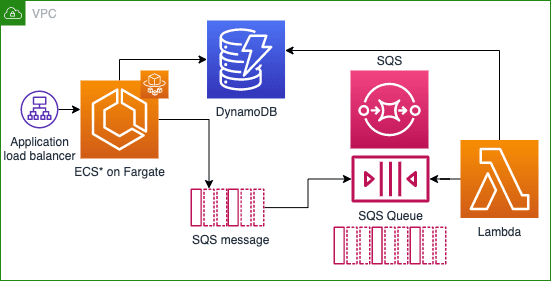
One of the projects I’ve been working on this summer has involved building a microservice called Strand that anneals DNA (joins two complementary strands). Users (mainly Organism Engineers) would use this technology to digitally replicate and verify the reactions they perform in the lab. The logic to perform the function already exists, but we want to separate it from its current home in a monolithic app that Ginkgo developed called OrganiCK, and implement a new architecture to handle requests to the service. This change will help make improvements on the current solution, including making anneal logic usable outside OrganiCK and reducing unnecessary complexity in the design.
This project is especially exciting to me for two reasons– firstly, because the final product will help alleviate scalability, resilience and stability concerns with an important tool, and secondly because I get to work with the AWS CDK, which is a new and powerful framework for defining infrastructure as code as Python modules.
In the first part of my project, I used CDK to create an ECS Fargate cluster, an API gateway to respond to requests, a Lambda to perform Strand logic and return outcomes, an SQS queue to respond to requests, and a DynamoDB instance to store results.
This fall, I will be connecting the DNA annealing logic with this infrastructure, but currently I’m working on a project that is supporting Ginkgo’s efforts to scale COVID-19 testing to millions of samples per day. It’s definitely exciting to be working on something that will have a positive impact, and it’s also been interesting to see how the planning and prototyping of new projects works in an industry setting.
Working with my mentor and other teammates and using technology I hadn’t encountered before have been my favorite parts of my internship so far, and I can’t wait to learn more as I continue to work on the Strand and COVID projects!
As the summer begins to wind down, we want to thank our six interns for their contributions to the Digital Tech Team, Ginkgo, and making biology easier to engineer. What they have accomplished this summer has been impressive. So, thank you, and we wish you great success in all the things that you will be doing next!
(Feature photo by Ramon Kagie on Unsplash)
Posted by Joseph Min, Addie Petersen, and Vidya Raghvendra