Ginkgo.Your Innovation Partner.

The Digital Technology Team welcomed six interns this summer. This is the first of a three-part series where our interns tell you about their summer in their own words. We will begin with Vichka and Etowah!
Hello! My name is Vichka Fonarev and I am a rising senior studying Computer Science at Tufts University. I have always been fascinated by the intersection of software engineering and the physical world so I was thrilled to join Ginkgo Bioworks’ Infragon, a team that works at the intersection of biology, software, and automation.
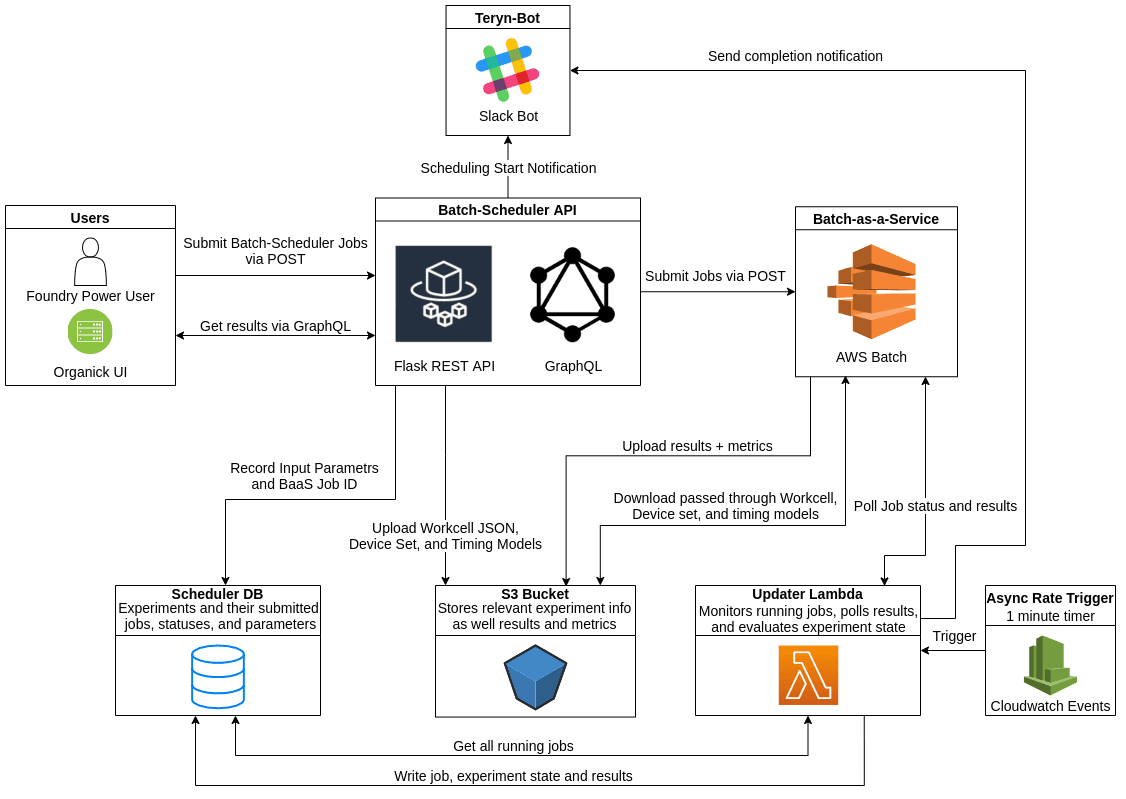
My project this summer was to implement a new architecture for the Batch Scheduler, a system that converts and ‘schedules’ Autoprotocol to a series of instructions that our workcells can understand and execute. In Ginkgo’s Foundry, we use a constraint solver from Strateos called SCLE to do this. However, many of Ginkgo’s Autoprotocol workflows are very complex and difficult to schedule. To combat this, a member of the NGS team wrote a tool called the Batch Scheduler. This tool allowed users to input many different constraints — such as batch sizes and overall scheduling time limitations — and would then generate all possible permutations of these constraints. This ‘batch’ of jobs could then be individually scheduled to increase the chance of getting a fast run, or at least a successful one.
While this worked, it was prone to failure and had load limitations due to a reliance on the manual setup of Celery workers. The re-architecture aimed to resolve both of these issues. The primary change in the re-architected system was moving the computationally intensive constraint solving from celery workers to Batch-as-a-Service (BaaS). BaaS is an internal tool that provides an abstraction on top of AWS Batch that makes setting up the infrastructure and submitting jobs significantly easier. The move to BaaS posed an interesting challenge since it is a closed asynchronous system where no results are directly returned and there is no completion notification. I had to implement an asynchronous lambda function that would poll BaaS jobs and write results to our database as they become available. Another big benefit of this new architecture is that we are now able to access the results of individual scheduling runs as they finish rather than waiting for the entire batch to finish. This allows scientists to run a workable schedule as soon as one becomes available rather than waiting for marginally more efficient schedules to be computed. Since we now rely on Batch job definitions rather than hosted celery servers, we are able to automatically deploy them from our CI/CD pipeline with no downtime for users. Additionally, we can now support multiple SCLE code versions so Foundry users who required different features can work concurrently on the same system.
Over the summer Ginkgo helped us integrate into the company culture and helped us meet as many people as possible. One of my favorite activities was a weekly Ask Me Anything (AMA) session with senior software developers set up by David Hofer, another software engineer. These AMAs were a great way to get some face-time with engineers from all sorts of backgrounds and from the diverse set of teams at Ginkgo. We got a ton of fantastic career advice and got to learn about many of the projects happening on the software team.
I also gained valuable experience working on a system with many moving parts. I learned not only a suite of software tools, but also how to tie together different services into a complex system in a way that’s efficient, scalable, and easy to understand.
Hi I’m Etowah! I’m a junior at Yale University where I study biology and computer science. This summer I worked as a software intern at Ginkgo Bioworks. It was a great experience and I learned a tremendous amount about building software and the challenges of doing biology at the scale that Ginkgo is doing it. I also made some close friends along the way.
One of the most prominent questions on my mind this summer was “how will Ginkgo standardize processes in biological engineering?”
I wondered about this particularly because I was coming to Ginkgo after spending the semester doing wet lab research in my molecular biology professor’s lab. The work I did there was extremely interesting, and different in nature to the work done at Ginkgo. Whereas the research at my professor’s lab does not require a great deal of automation and process standardization, Ginkgo aims to have many more projects in flight than individuals working directly on them, and with significant divisions of labor.
Standardization is one of the keys that allows Ginkgo to scale the number of its projects. Standardizing operations and information enables efficient processes and handoffs between teams. But how could something as complex as engineering in biology be standardized?
Through my internship, it has become clear to me that at Ginkgo, software will play a central role in creating and fostering these standardizations. In my time here, I worked on helping promote two types of standardizations: the first software-oriented, and the second more biology-oriented.
Ginkgo has many different user-facing web interfaces, many of which are written in React, a Javascript frontend web framework. These interfaces, such as for the inventory or workflow builder, were largely developed independently of each other so each element of each interface is unique. Such being the case, it is harder to transfer knowledge of one interface to another.
In my first project, I created a React component library and published it to Ginkgo’s internal node package manager to allow these interfaces to use the same components. Using standard components across interfaces reduces development time and decreases the amount of time it takes scientists to learn to use them, allowing them to focus on the biology.
My second project was born in response to a new centralized strain banking service. Rather than being stored in miscellaneous freezers, important microbial strains would be stored in a centralized location. Banked strains are easier to find and can be reused more readily across projects.
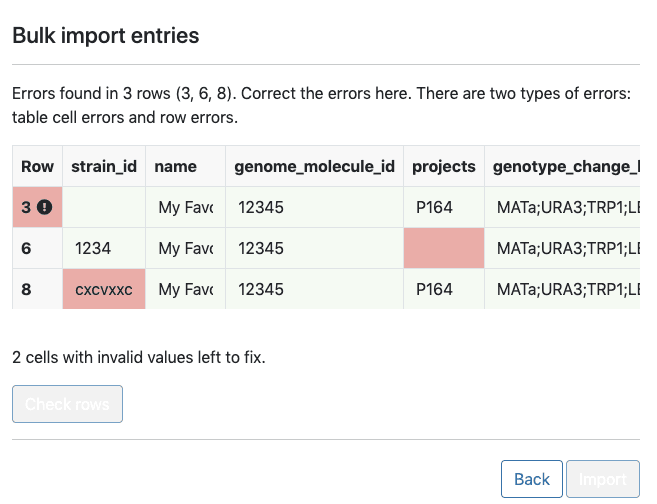
Standardized information about each banked strain, such as strain origin and usage, must also be recorded in Ginkgo’s laboratory inventory management system (LIMS). To do this, scientists upload a CSV file containing metadata for many strains to LIMS.
If the CSV file is incorrectly formatted or is missing fields, though, they must edit the CSV and upload it again. I sought to allow scientists to seamlessly upload and correct these errors without having to return to the CSV file itself. I did this by creating a React component that detects and fixes these issues. This makes the process of creating strains with standardized metadata less time consuming. I then deployed a React application with this component (to an autoscaling serverless stack I stood up on AWS).
As Ginkgo continues to grow, enabling standardization of biological engineering practices through software is a must. The projects I worked on this summer contribute to this. In all, I had a fantastic summer and am excited to have contributed to Ginkgo’s mission of making biology easier to engineer.
Stay tuned! In our next blog post, we will be hearing about the cool things that Liam and Aileen have been doing this summer!
(Feature photo by frank mckenna on Unsplash)
Posted by Etowah Adams and Vichka Fornarev