Ginkgo.Your Innovation Partner.
At Ginkgo, our mission to make biology easier to engineer constantly challenges our software team to develop innovative solutions that drive us forward. One of the questions we have focused on recently is ‘How can we make the process of designing DNA more efficient?’
To address this challenge, our new DNA design platform was built with the goal of both enabling large scale design and quick iterations on biological hypotheses. However, to keep up with Ginkgo’s pace of scaling the design of new organisms, we need to consider all software solutions, including leveraging open source technologies. In this instance, Lattice Automation’s open source sequence viewer, SeqViz, was an excellent option for our users’ needs and accelerating our development timeline.
The combination of these parts often results in DNA libraries of tens of thousands of constructs. Our DNA tool streamlines this process by enabling users to access and manage a curated set of reusable genetic parts. These parts can easily be combined and a single-click mechanism enables ordering the synthesis of the resulting libraries.
Because of the huge number of constructs per library, biologists need to quickly spot-check their work to ensure the resulting sequences are free of errors or unexpected genetic elements. This is best accomplished with a sequence visualizer that clearly illustrates designed plasmids, their nucleotide sequences, and clear annotations of genetic features.
Sequence viewers are not trivial to build, and given the numerous enterprise options available in the industry, most biologists have a good idea of what to expect from them. Developing a brand new sequence viewer would have required significant effort and would have prevented us from focusing on our goal of augmenting Ginkgo’s cutting edge synthetic biology workflows with software. Therefore, we turned to the open source community in hopes of finding a suitable solution.
[caption id="attachment_3991" align="aligncenter" width="2872"]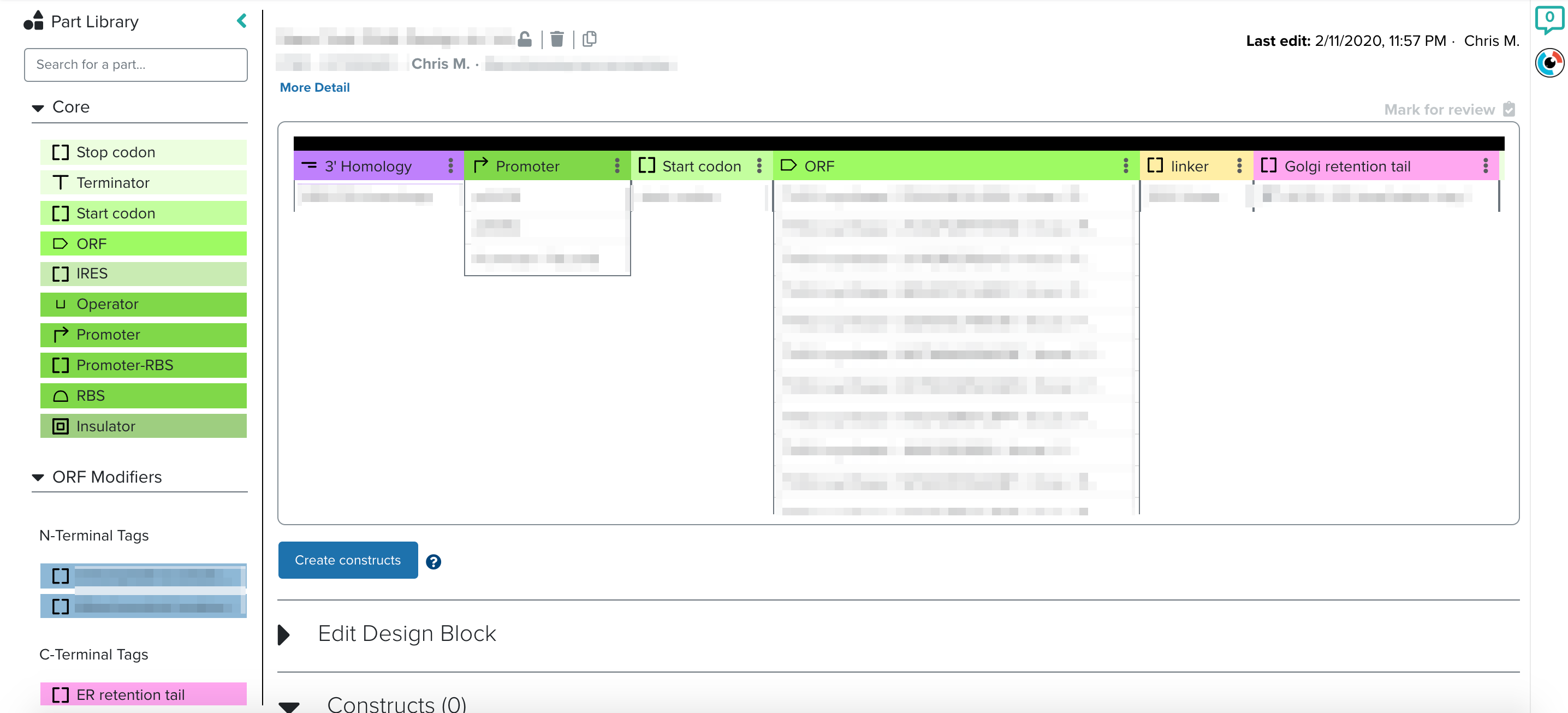 The challenge with building our new DNA Design: enabling both large scale design and quick iteration[/caption]
The challenge with building our new DNA Design: enabling both large scale design and quick iteration[/caption]
Ginkgo’s unique, cutting-edge environment presents us with a wide range of software use cases that will help us to contribute meaningful feedback to new and innovative projects like SeqViz, and we are excited to collaborate on future improvements. Lattice has already incorporated our feedback by adding features like 6-frame translation. Although this addition was requested by Ginkgo, we are confident it will benefit many other users by making it easier to verify the presence and validity of coding sequences (CDS) and ensure that unwanted genes are not introduced in a design.
[caption id="attachment_4010" align="alignnone" width="2876"]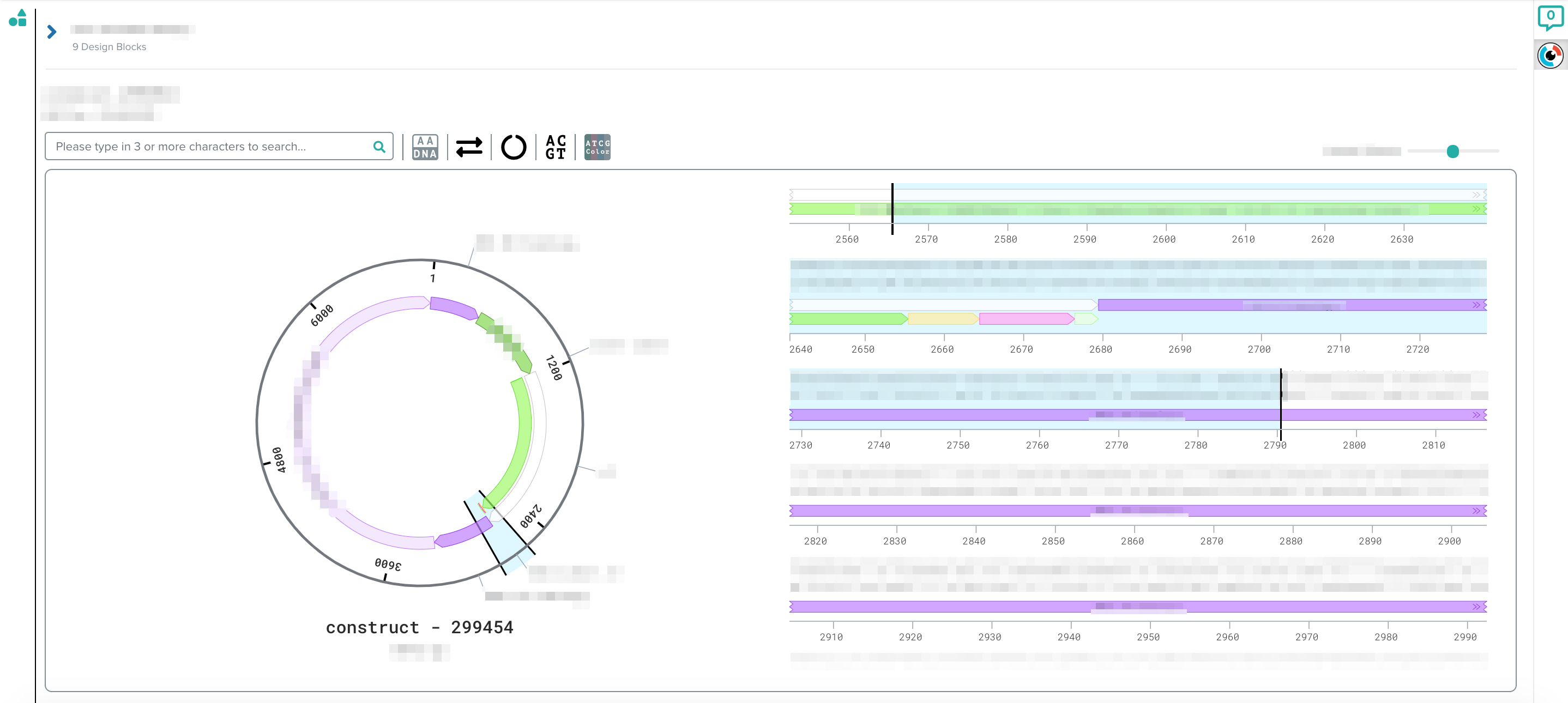 SeqViz provides a rich view on the genetic parts and designed constructs in our DNA design tool.[/caption]
SeqViz provides a rich view on the genetic parts and designed constructs in our DNA design tool.[/caption]
Powerful software tools reduce the overhead associated with designing, iterating through, and analyzing hypotheses. This efficiency in turn helps scientists use Ginkgo’s genetic engineering platform to rapidly prototype biology. By continuing to engage with open source projects, we can help make open source software tools more robust and reusable. Ginkgo and other innovators in our space can then focus less on developing their software tool kit and more on solving complex biological problems. This will ultimately reduce the cost and time needed to make meaningful impact with biological engineering.
Posted by Chris Murphy