Ginkgo.Your Innovation Partner.

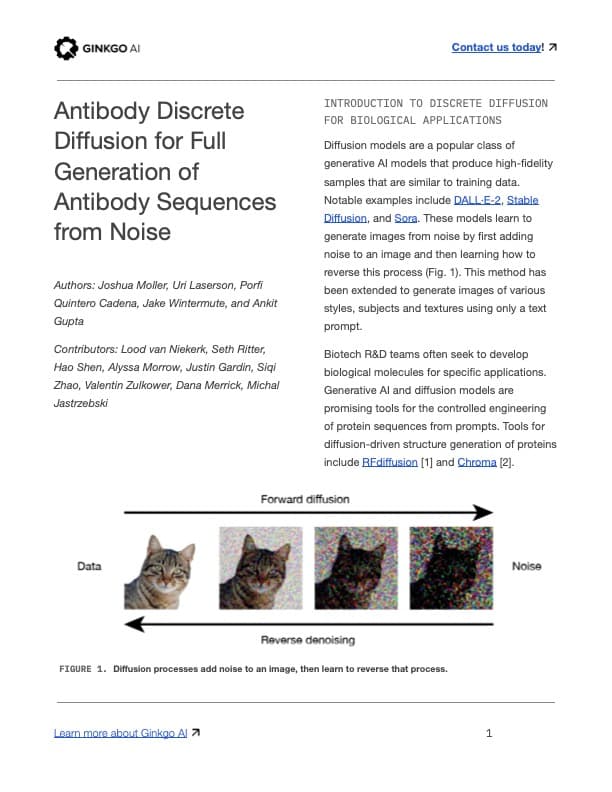
Diffusion models are a popular class of generative AI models that produce high-fidelity samples that are similar to training data. Notable examples include DALL·E-2, Stable Diffusion, and Sora. These models learn to generate images from noise by first adding noise to an image and then learning how to reverse this process (Fig. 1). This method has been extended to generate images of various styles, subjects and textures using only a text prompt.
Biotech R&D teams often seek to develop biological molecules for specific applications. Generative AI and diffusion models are promising tools for the controlled engineering of protein sequences from prompts. Tools for diffusion-driven structure generation of proteins include RFdiffusion [1] and Chroma [2].
Structure-focused generation offers a promising tool for many design applications, such as designing and predicting binding to target proteins. However, its reliance on well-defined structures complicates its use for applications where the critical structural intermediate is unknown. Biological sequence data may help to fill this gap.